Researchers from LSTM have now completed an in-depth analysis of the genomes for two Wolbachia bacterial endosymbiont strains that infect Anopheles mosquitoes. Their work, published in Microbial Genomics, shows significantly reduced prophage regions in both strains compared to previously sequenced Wolbachia genomes, with potential implications on how these strains affect their Anopheles hosts.
As part of a large international team including scientists from Cameroon, the DRC and the UK including LSTM’s mosquito microbiome researchers Dr. Grant Hughes and Dr. Eva Heinz had previously published the first strong evidence for a high density, natural infection of Anopheles mosquitoes with two different strains of Wolbachia. This discovery had opened the door for potential mosquito control strategies for malaria in other Anopheles species.
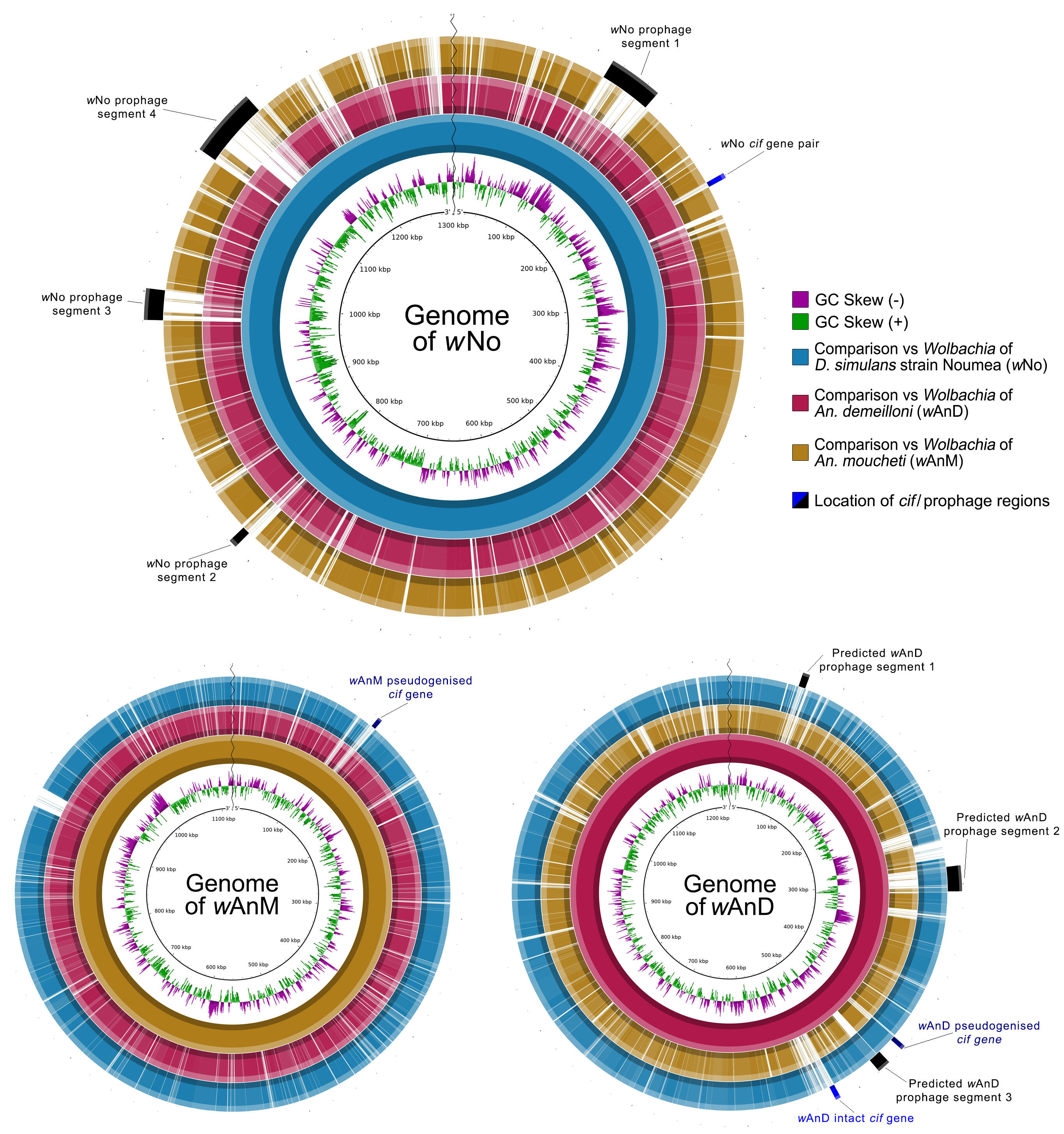
The researchers now present an in-depth analysis of the genomes for these Anopheles-associated Wolbachia strains, led by Dr. Shannon Quek. This showed that, while both strains had reduced genome sizes compared to previously sequenced Wolbachia, they still contained metabolic pathways commonly associated with Wolbachia-host interactions. Instead, the streamlining of the genome can be attributed to reduced prophage elements, with one strain lacking them almost entirely.
Prophages are genomic segments of bacteriophages that have integrated themselves into the bacterial genome. Such mobile genetic elements often move horizontally between different bacteria and can encode for genes that allow adaptation to specific environments, such as virulence factors in pathogenic bacteria. In Wolbachia, these regions are often known to contain genes that manipulate host reproductive success, such as the relatively-recently characterised Cytoplasmic Incompatibility Factor (cif) genes.
Dr. Quek, the study’s first author, explains: “The significantly reduced size of such regions within these two Wolbachia strains is of interest, particularly when viewed in context of their high density and high prevalence rates, in naturally infected Anopheles populations. Furthermore, while we were able to identify cif genes within the genomes of both strains, only one genome contained a fully intact copy. How a strain persists in a population without functional cif genes remains unknown, but we have an opportunity to investigate that in these mosquitoes. Regardless, both strains hold great promise for applied vector control approaches aimed at curbing malaria.”
Dr. Heinz, the senior author of the study: “One of the interesting observations is that these two Wolbachia lineages seem to have been acquired by the two Anopheles species independently. This, in addition to the heterogeneous infection of both host populations with Wolbachia as shown in our earlier work, opens exciting avenues to understand what factors make Anopheles permissive or impede a symbiosis with Wolbachia. To fully resolve their evolutionary history and address these questions, however, we need more data from the host and associated bacteria, to understand the larger population dynamics and genomic diversity of the host and the bacterium and their co-evolutionary history.”
Shannon Quek, Louise Cerdeira, Claire L. Jeffries, Sean Tomlinson, Thomas Walker, Grant L. Hughes and Eva Heinz
Wolbachia endosymbionts in two Anopheles species indicates independent acquisitions and lack of prophage elements
Microbial Genomics 8, Apr 2022 https://doi.org/10.1099/mgen.0.000805