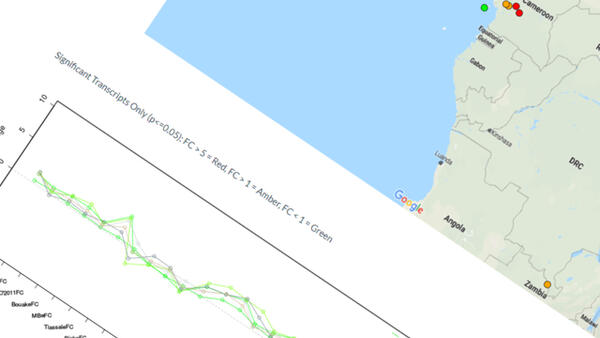
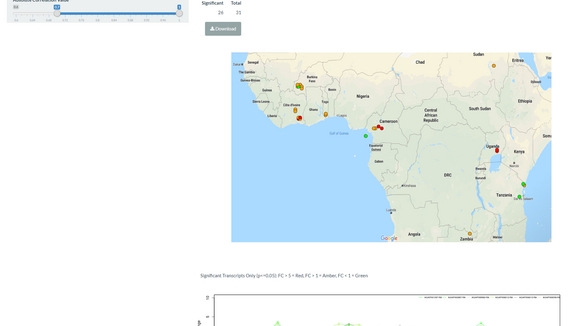
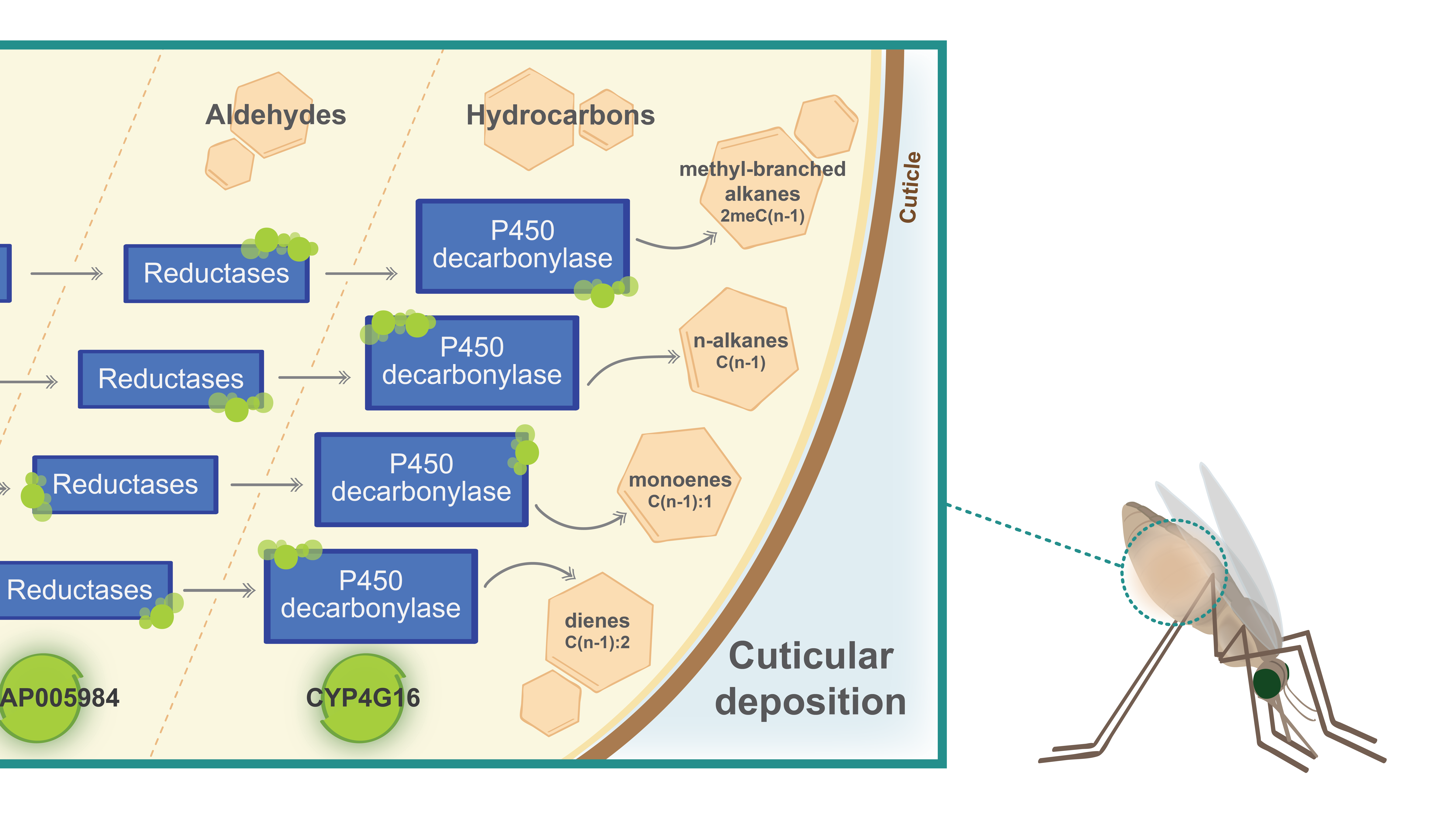
The unit develops and deploys bioinformatics tools to translational tropical medicine and is engaged in a number of collaborative projects with other research groups in LSTM.
LSTM’s Bioinformatics Unit activities, led by Dr Simon Wagstaff, have entered a new software development phase as it explores novel ways to address some of the major computational challenges that limit our ability to analyse the results of high throughput sequencing projects. To address these unmet needs the unit developed VTBuilder, a new tool for the assembly of highly variable neglected transcriptomes from millions of short sequencing reads. This substantial piece of software is now being used in the BU to accurately assemble highly variable transcriptomes from non-model organisms lacking a comprehensive underpinning knowledge and technology base.
Consistent with the software development theme, Dr Wagstaff has been working closely with staff at the NERC Bioinformatics Analysis Facility in Wallingford (NBAF-W) to integrate Galaxy software into Biolinux 8. This workflow-based environment offers a more productive development platform for local users to experiment with complicated pipelines and has already been used to identify over 200 new genes in Wolbachia ‘missing’ from the original annotated genome.