
ADDovenom
Novel Snakebite Therapy Platform of Unparalleled Efficacy, Safety and Affordability


FET OPEN scheme, funded by the EC, H2020 research programme
ADDovenom initiates a major multidisciplinary effort to achieve a timely step-change in snakebite therapy, by creating an innovative platform with transformative potential for antivenom generation to save countless lives.
ADDovenom exploits a disruptive new, protein-based nanoscaffold we developed, the ADDomer - a megadalton sized, thermostable synthetic virus-like particle that offers 60 high-affinity binding sites to rapidly eliminate venom toxins from the blood stream. Further, we will for the first time deploy ADDobody, a small, stable protein motif with randomized flexible loops that will be utilized as a naïve library to select and evolve high-affinity binders in vitro by Ribosome Display. Of note, the ADDobody and ADDomer ‘superbinder’ formats are interconvertible.
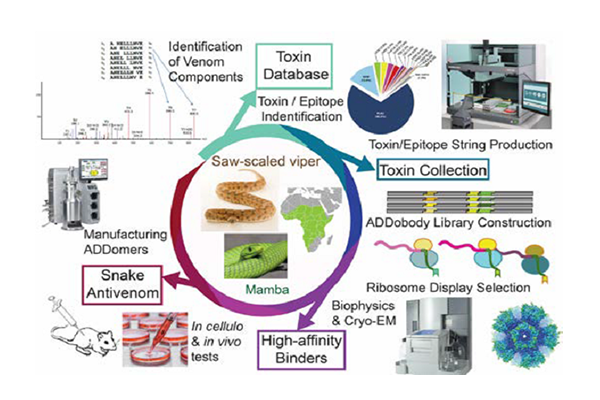
The ADDovenom project brings to bear cutting-edge proteomics, transcriptomics and bioinformatics to inventories the toxin repertoire of eight snakes that inflict the most clinically-challenging envenoming syndromes in sub-Saharan Africa: haemorrhage, coagulopathy, paralysis and tissue necrosis. We will implement rational design and highthroughput expression to produce antigens for our selections, based on the major toxin groups we target. We will design consensus-toxins and epitope strings combining conserved sequences, to achieve maximal intergeneric efficacy of our ADDobody binders, boosting neutralizing efficacy for entire toxin families simultaneously. We will develop state-of-the-art bioprocessing to manufacture ADDomer-based antivenoms at pharma scale, preparing for future clinical trials.
To achieve these ambitious goals, ADDovenom synergistically combines unique expertise across a range of techniques and scientific disciplines, towards the objective to develop easy to produce, first-in-class neutralizing superbinders for snakebite therapy, to protect with unprecedented efficacy against the most prevalent snakebites – a strategy that can be adapted to all snakes in any geographic region.
What we do
The Challenge: The first-choice treatment of snakebite envenoming is antivenom and yet this Neglected Tropical Disease still, annually, causes up to 138,000 deaths and 400,000 disabilities in surviving victims. This disease burden primarily affects the most economically and medically disadvantaged farming communities of Asia, sub-Saharan Africa and Latin America. Current anti-venoms (AVs) are based on antibodies from hyperimmunised horses and sheep; an approach implemented in 1896 by Albert Calmette. These AVs are weakly effective: only 10-15% of the IgGs bind venom, and multiple vials are needed to effect cure, but each additional vial induces higher levels of adverse effects and increases treatment costs. This approach has many additional disadvantages, in particular it cannot rationally incorporate the distinct immunogenicity or toxicity of the venoms’ proteins into the design(venoms comprise between 20 to >100 proteins that vary in molecular mass, bioactivity and pathogenicity). This is particularly problematic because snake venoms and their toxins vary significantly at every taxonomic level, and highly pathogenic, diverse and low-molecular mass toxins are notoriously poor immunogens.

An innovative solution requiring proof-of-principle validation
We propose a radical, innovative change to snakebite treatment by rationally designing ADDobodies/ADDomers (ADDovenom) to bind and neutralise the pathogenic function of all toxins of the medically most important snakes, irrespective of taxonomy or geography, and without adverse effect risk. We will achieve this by applying cutting-edge analysis techniques to snake venoms to enable informed target choices, by establishing new protein engineering tools, and high-throughput production methods. Based on non-conventional techniques and designs, new protein-based nanoparticles - ADDobodies and ADDomers - will be implemented with cost-effective technologies and compatible with industrial processes and quality control. The latter will be developed in collaboration with the non-academic partner towards a rapid industrial roll-out and downstream (pre-)clinical trials.
Implementation of the project
The project comprises several technological challenges and high-risk research activities.
The scientific work is organised in four highly interdependent Work Packages (WP2-WP5), led by the Principal Investigators and carried out by PRDAs with PhDs in relevant areas.
WP2 (Mass spectrometry and Bioinformatics) involves proteomics based in-depth venom characterization, followed by ‘omics’- and bioinformatics-based choice of target toxins/epitopes and design of antigens.
WP3 (In vitro evolution and characterization of ADDobody binders) will produce these rationally designed antigens and use them for in vitro selection and evolution of ADDomers using a naïve ADDobody library.
WP3 and WP4 (Examination of neutralizing ability) will analyse the selected ADDobodies and ADDomer superbinders in vitro and in vivo for affinity, specificity and neutralizing ability.
WP5 (Scalable bioprocess for ADDomer production) develops protocols for intensified, cost-efficient ADDomer manufacture compatible with industrial processes.
Results from WP3 and WP4 characterizations will feed-back directly into WP2 and WP3 to refine design considerations for antigens and ADDobodies as well as Ribosome Display selection strategies.
Optimised protocols from WP5 will be fed back to the production of ADDomers for structural characterization in WP3. WP4 findings may also influence the manufacturing process (WP5). In addition, all results will be channelled through the PIs, who as well as managing the work packages, will provide the vision to steer the direction of the project.
Finally WP1 (Project Management) will manage the project in terms of compliance and reporting to the EC, ensure deliverables and milestones are met, and coordinate WP6 (Communication, dissemination and exploitation of results).

University of Bristol (Lead) will express, purify and characterise ADDobodies and ADDomers, generate a naïve ADDobody library, carry out in vitro selection/evolution by Ribosome Display, provide sample for in cellulo and in vivo characterisation, constructs for manufacturing antivenoms and determine structures.

University of Liege will be in charge of analysing the venoms and sequencing their toxins by various proteomics approaches, with the aim of sequencing the most toxic toxins, whatever their size or their abundancy in the venom.

Liverpool School of Tropical Medicine will provide venoms and venom gland transcriptomes for eight snakes, basic “serology” of ADDomer recognition of toxin components, determination of the ability of ADDomers and ADDobodies to in vitro neutralise the function of each class of toxins and comprehensive pre-clinical in vivo testing of the best ADDomer candidates in murine models of venom lethality. Further, LSTM will perform pharmacokinetic characterizations of ADDomers in rodent models and examination of toxicity of ADDomers in a rodent model. LSTM will advocate for the next phase of ADDovenom research – clinical trial preparedness in sub-Saharan Africa.

Universite D’Aix Marseille will be in charge of the expression and purification of most of the toxins, epitope strings, ADDobodies and ADDomers of the project. Purified proteins and complexes will be characterised biochemically and biophysically by several methods including ELISA, surface plasmon resonance (SPR), Thermophoresis, SECMALS and thermal shift assays

Instituto de Biologica Experimental e Tecnologica will work on the implementation of supporting analytical methods; upstream and downstream process development; production of ADDomers.